Reproducible Research Data Management with DataLad
6th RDM-Workshop 2024 on Research Data Management in the Max Planck Society
March 20, 2024
About

Dr. Lennart Wittkuhn
wittkuhn@mpib-berlin.mpg.de
https://lennartwittkuhn.com/
Mastodon GitHub LinkedIn
About me
I am a Postdoctoral Research Data Scientist in Cognitive Neuroscience at the Institute of Psychology at the University of Hamburg (PI: Nicolas Schuck)
BSc Psychology & MSc Cognitive Neuroscience (TU Dresden), PhD Cognitive Neuroscience (Max Planck Institute for Human Development)
I study the role of fast neural memory reactivation in the human brain, applying machine learning and computational modeling to fMRI data
I am passionate about computational reproducibility, research data management, open science and tools that improve the scientific workflow
Find out more about my work on my website, Google Scholar and ORCiD
About this presentation
Slides: https://lennartwittkuhn.com/talk-mps-fdm-2024
Source: https://github.com/lnnrtwttkhn/talk-mps-fdm-2024
Software: Reproducible slides built with Quarto and deployed to GitHub Pages using GitHub Actions for continuous integration & deployment
License: Creative Commons Attribution-ShareAlike 4.0 (CC BY-SA 4.0)
Contact: Feedback or suggestions via email or GitHub issues. Thank you!
Acknowledgements and further reading
Slides and presentations by Dr. Adina Wagner and the DataLad team, e.g., “DataLad - Decentralized Management of Digital Objects for Open Science” (Wagner 2024)
The DataLad Handbook by Wagner et al. (2022) is a comprehensive educational resource for data management with DataLad.
Papers
- Wilson et al. (2014). Best practices for scientific computing. PLOS Biology.
- Wilson et al. (2017). Good enough practices in scientific computing. PLOS Computational Biology.
- Lowndes et al. (2017). Our path to better science in less time using open data science tools. Nature Ecology Evolution.
Talks
- Richard McElreath (2020). Science as amateur software development. YouTube
- Russ Poldrack (2020). Toward a Culture of Computational Reproducibility. YouTube
… and many more!
Agenda
1. Scientific workflows with DataLad
1.1 Version Control
1.2 Modularity & Linking
1.3 Provenance
1.4 Collaboration & Interoperability
2. Integrating DataLad with MPS infrastructure (and beyond)
2.1 Keeper
2.2 Edmond
2.3 ownCloud / nextCloud
2.4 GIN
2.5 GitLab
3. Summary & Discussion
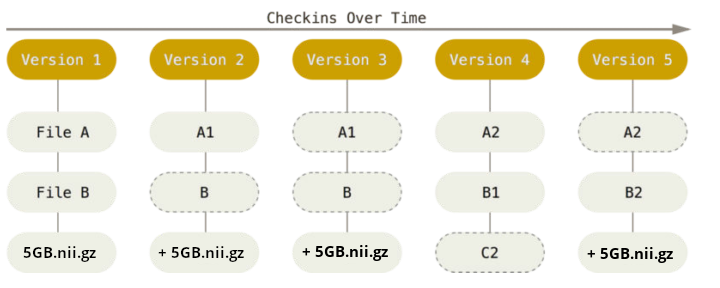
Scientific building blocks are not static
We need version control
Why we need version control
… for code (text files) 
… for data (binary files) 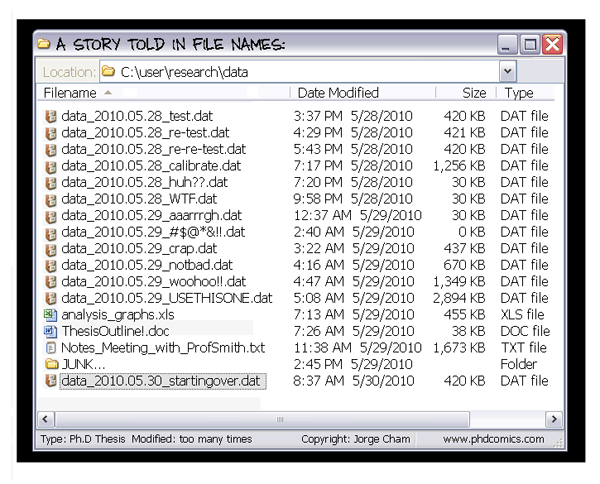
If everything is relevant, track everything.
What is version control?
“Version control is a systematic approach to record changes made in a […] set of files, over time. This allows you and your collaborators to track the history, see what changed, and recall specific versions later […]” (Turing Way)
keep track of changes in a directory (a “repository”)
take snapshots (“commits”) of your repo at any time
know the history: what was changed when by whom
compare commits and go back to any previous state
work on parallel “branches” & flexibly “merge” them
“push” your repo to a “remote” location & share it
share repos on platforms like GitHub or GitLab
work together on the same files at the same time
others can read, copy, edit and suggest changes
make your repo public and openly share your work
What are Git and DataLad?
- most popular version control system
- free, open-source command-line tool
- graphical user interfaces exist, e.g., GitKraken
- standard tool in the software industry
- 100 million GitHub users 1
Sadly, Git does not handle large files well.
- “Git for (large) data”
- free, open-source command-line tool
- builds on top of Git and git-annex
- allows to version control arbitrarily large datasets 2
- DataLad Python API: Use DataLad in your Python code
- graphical user interface exists: DataLad Gooey
Example Dataset: Brain Imaging Data
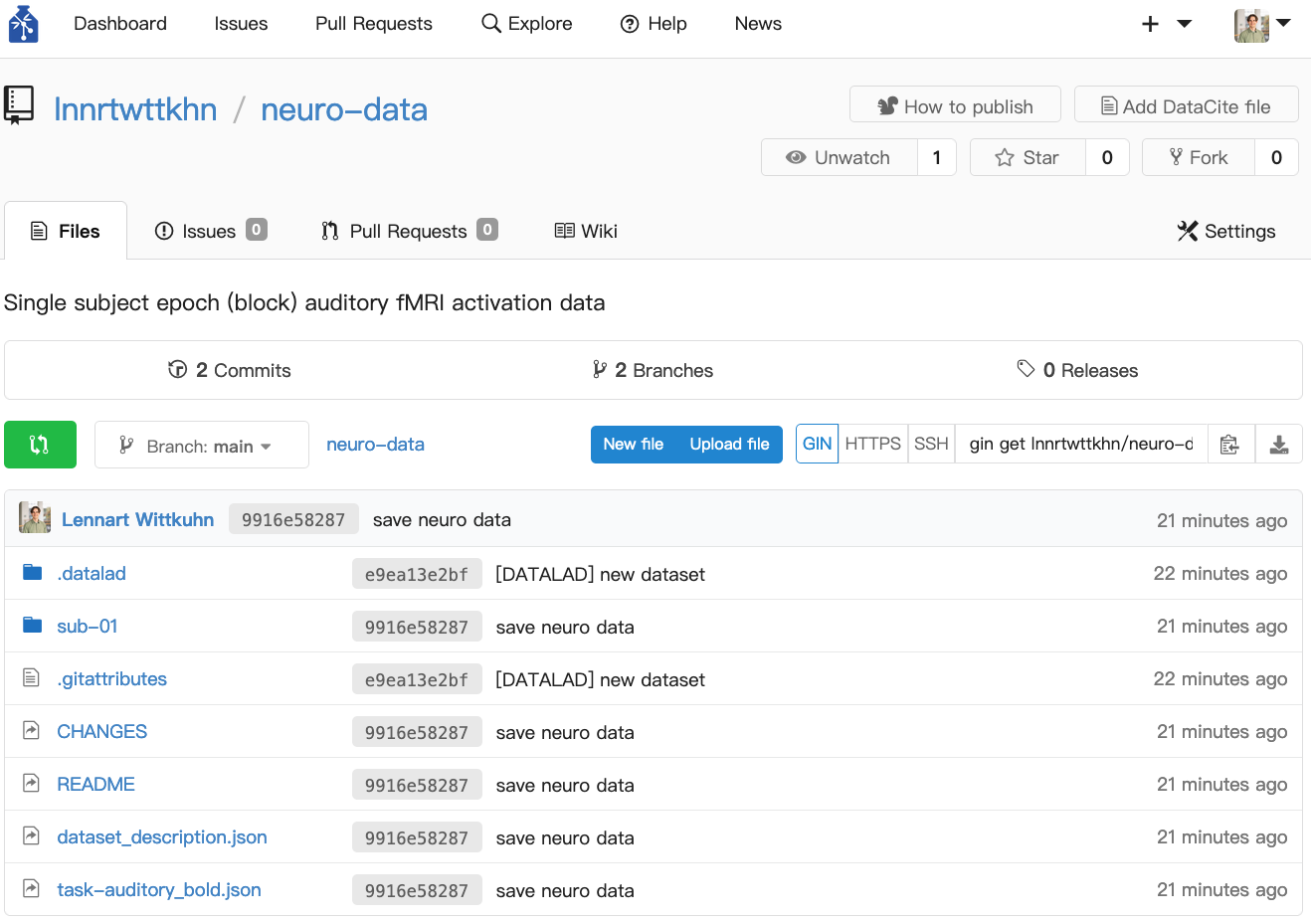
Single subject epoch (block) auditory fMRI activation data
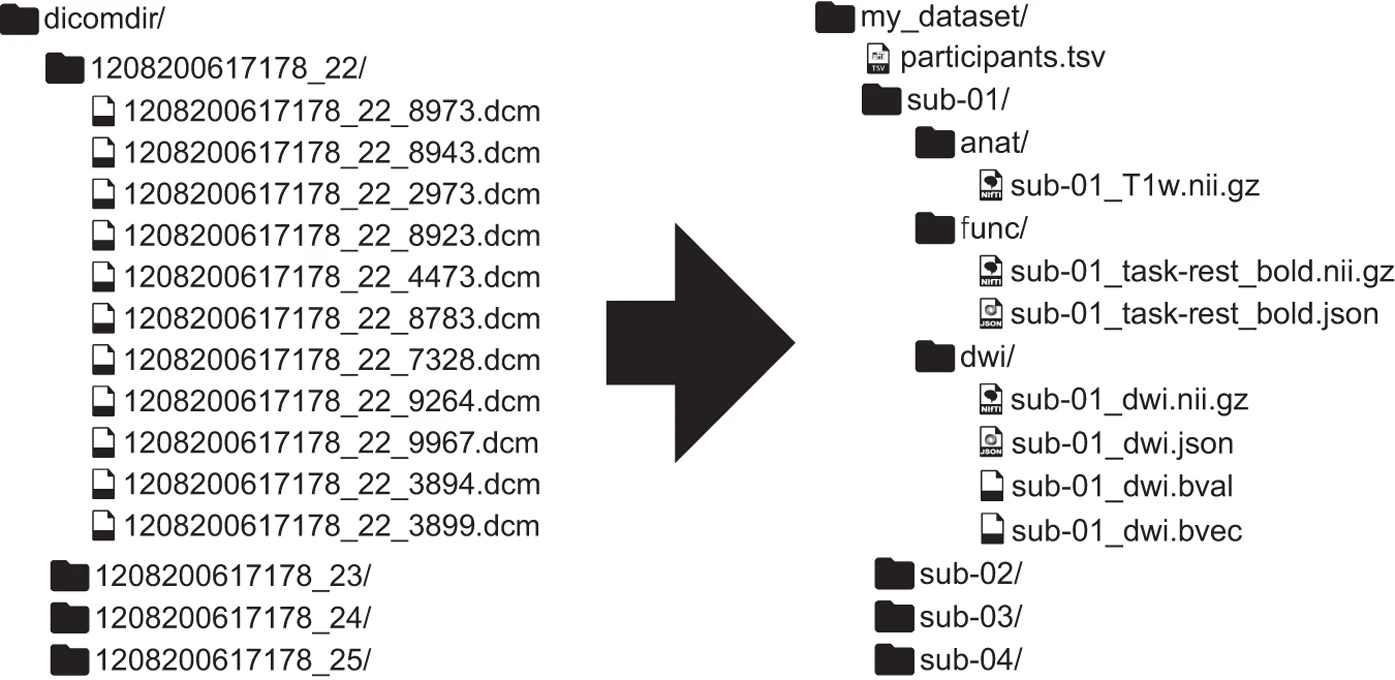
Dataset from Functional Imaging Laboratory, UCL Queen Square Institute of Neurology, London, UK (Source)
Version Control with DataLad
View output
add(ok): CHANGES (file)
add(ok): README (file)
add(ok): dataset_description.json (file)
add(ok): sub-01/anat/sub-01_T1w.nii (file)
add(ok): sub-01/func/sub-01_task-auditory_bold.nii (file)
add(ok): sub-01/func/sub-01_task-auditory_events.tsv (file)
add(ok): task-auditory_bold.json (file)
save(ok): . (dataset) action summary:
add (ok: 7)
save (ok: 1)Data in DataLad datasets are either stored in Git or git-annex
Git
- handles small files well (text, code)
- file contents are in Git history and will be shared
- Shared with every dataset clone
- Useful: small, non-binary, frequently modified files
git-annex
- handles all types and sizes of files well
- file contents are in the annex, not necessarily shared
- Can be kept private on a per-file level
- Useful: Large files, private files
Science is build from modular units
We need modularity and linking
Version control beyond single repositories
Research as a sequence
- Prior works (code development, empirical data, etc.) are combined to produce results with goal of a publication
- Aggregation across time and contributors
- Aiming for (but often failing) to be reproducible
- Often, there is one big project folder
A single repository is not enough!
Nesting of modular DataLad datasets
- seamless nesting of modular datasets in hierarchical super-/sub-dataset relationships
- based in Git submodules, but mono-repo feel thanks to recursive operations
- overcomes scaling issues with large amounts of files (Example: Human Connectome Project)
- modularizes research components for transparency, reuse and access management
Example: Intuitive data analysis structure
First, let’s create a new data analysis dataset:
datalad create -c yoda myanalysis
[INFO ] Creating a new annex repo at /tmp/myanalysis
[INFO ] Scanning for unlocked files (this may take some time)
[INFO ] Running procedure cfg_yoda
[INFO ] == Command start (output follows) =====
[INFO ] == Command exit (modification check follows) =====
create(ok): /tmp/myanalysis (dataset)-c yoda initializes useful structure (details here):
We install analysis input data as a subdataset to the dataset:
datalad clone -d . https://github.com/datalad-handbook/iris_data.git input/
[INFO ] Remote origin not usable by git-annex; setting annex-ignore
install(ok): input (dataset)
add(ok): input (dataset)
add(ok): .gitmodules (file)
save(ok): . (dataset)
action summary:
add (ok: 2)
install (ok: 1)
save (ok: 1)Modular units with clear provenance
git diff HEAD~1
diff --git a/.gitmodules b/.gitmodules
new file mode 100644
index 0000000..fc69c84
--- /dev/null
+++ b/.gitmodules
@@ -0,0 +1,5 @@
+[submodule "input"]
+ path = input
+ url = https://github.com/datalad-handbook/iris_data.git
+ datalad-id = 5800e71c-09f9-11ea-98f1-e86a64c8054c
+ datalad-url = https://github.com/datalad-handbook/iris_data.git
diff --git a/input b/input
new file mode 160000
index 0000000..b9eb768
--- /dev/null
+++ b/input
@@ -0,0 +1 @@
+Subproject commit b9eb768c145e4a253d619d2c8285e540869d2021Science is exploratory and iterative
We need provenance
Reusing previous work is hard
Establishing provenance with DataLad
datalad run wraps around anything expressed in a command line call and saves the dataset modifications resulting from the execution.
datalad rerun repeats captured executions. If the outcomes differ, it saves a new state of them.
datalad containers-run executes command line calls inside a tracked software container and saves the dataset modifications resulting from the execution.
datalad containers-run \
--message "Time series extraction from Locus Coeruleus"
--container-name nilearn \
--input 'mri/*_bold.nii' \
--output 'sub-*/LC_timeseries_run-*.csv' \
"python3 code/extract_lc_timeseries.py"
-- Git commit --
commit 5a7565a640ff6de67e07292a26bf272f1ee4b00e
Author: Adina Wagner adina.wagner@t-online.de
AuthorDate: Mon Nov 11 16:15:08 2019 +0100
[DATALAD RUNCMD] Time series extraction from Locus Coeruleus
=== Do not change lines below ===
{
"cmd": "singularity exec --bind {pwd} .datalad/environments/nilearn.simg bash..",
"dsid": "92ea1faa-632a-11e8-af29-a0369f7c647e",
"inputs": [
"mri/*.bold.nii.gz",
".datalad/environments/nilearn.simg"
],
"outputs": ["sub-*/LC_timeseries_run-*.csv"],
...
}
^^^ Do not change lines above ^^^- Enshrine the analysis in a script and record code execution together with input data, output files and software environment in the execution command
- Result: Machine readable record about which data, code and software produced a result how, when and why
- Use the unique identifier (hash) of the execution record to have a machine recompute and verify past work
datalad rerun 5a7565a640ff6de67
[INFO ] run commit 5a7565a640ff6de67; (Time series extraction from Locus Coeruleus)
[INFO ] Making sure inputs are available (this may take some time)
get(ok): mri/sub-01_bold.nii (file)
[...]
[INFO ] == Command start (output follows) =====
[INFO ] == Command exit (modification check follows) =====
add(ok): sub-01/LC_timeseries_run-*.csv(file)Science is collaborative & distributed
We need interoperability & transport logistics
Data sharing and collaboration with DataLad
“I have a dataset on my computer.
How can I share it or collaborate on it?”
Challenge: Scientific workflows are idiosyncratic across institutions / departments / labs / any two scientists
Share data like code
- With DataLad, you can share data like you share code: As version-controlled datasets via repository hosting services
- DataLad datasets can be cloned, pushed and updated from and to a wide range of remote hosting services
Interoperability with a range of hosting services
DataLad is built to maximize interoperability and streamline routines across hosting services and storage technology
Separate content in Git vs. git-annex behind the scenes
- DataLad datasets are exposed via private or public repositories on a repository hosting service (e.g., GitLab or GitHub)
- Data can’t be stored in the repository hosting service but can be kept in almost any third party storage
- Publication dependencies automate interactions between both paces
Special cases
Repositories with annex support
- Easy: Only one remote repository
- Examples: GIN, GitLab with annex support
Special remotes with repositories
- Flexible: Full history or single snapshot
- Examples: DataLad-OSF
Have access to more data than you have disk-space
Cloned datasets are lean.
“Metadata” (file names, availability) are present …
… but no file content:
File contents can be retrieved on demand:
datalad get .
get(ok): CHANGES (file) [from origin...]
get(ok): README (file) [from origin...]
get(ok): dataset_description.json (file) [from origin...]
get(ok): sub-01/anat/sub-01_T1w.nii (file) [from origin...]
get(ok): sub-01/func/sub-01_task-auditory_bold.nii (file) [from origin...]
get(ok): sub-01/func/sub-01_task-auditory_events.tsv (file) [from origin...]
action summary:
get (ok: 6)Let’s check the dataset size again:
Drop file content that is not needed:
datalad drop .
drop(ok): CHANGES (file) [locking origin...]
drop(ok): README (file) [locking origin...]
drop(ok): dataset_description.json (file) [locking origin...]
drop(ok): sub-01/anat/sub-01_T1w.nii (file) [locking origin...]
drop(ok): sub-01/func/sub-01_task-auditory_bold.nii (file) [locking origin...]
drop(ok): sub-01/func/sub-01_task-auditory_events.tsv (file) [locking origin...]
drop(ok): . (directory)
action summary:
drop (ok: 7)When files are dropped, only “metadata” stays behind, and files can be re-obtained on demand.
Data sharing using DataLad and data infrastructure of the Max Planck Society
Sharing DataLad datasets via Keeper
“A free service for all Max Planck employees and project partners with more than 1TB of storage per user for your researchdata.”
✨ Features of Keeper ✨
- > 1 TB per Max Planck employee (and expandable):
- based on cloud-sharing service Seafile
- data hosted on MPS servers
- configurable as a DataLad special remote
Sharing DataLad datasets via Edmond
“Edmond is a research data repository for Max Planck researchers. It is the place to store completed datasets of research data with open access.”
✨ Features of Edmond ✨
- based on Dataverse, hosted on MPS servers
- use is free of charge
- no storage limitation (on datasets or individual files)
- flexible licensing
Two modes:
annex mode (default): non-human readable representation of the dataset that includes Git history and annexed data
filetree mode: human readable single snapshot of your dataset “as it currently is” that does not include history of annexed files (but Git history)
Sharing DataLad datasets via ownCloud / Nextcloud
DataLad NEXT extension allows to push / clone DataLad datasets to / from ownCloud & Nextcloud (via WebDAV)
ownCloud GWDG: “50 GByte default storage space per user; flexible increase possible upon request”
✨ Features of ownCloud and Nextcloud ✨
- data privacy compliant alternative to Google Drive, Dropbox, etc. (usually hosted on-site)
- provided by your institution, so free to use
- supports private and public repositories
- can be used together with external collaborators
- expose datasets for regular download without DataLad
Sharing DataLad datasets via GIN
“GIN is […] a web-accessible repository store of your data based on git and git-annex that you can access securely anywhere you desire while keeping your data in sync, backed up and easily accessible […]“
✨ Features of GIN ✨
- free to use and open-source (could be hosted within your institution; for more details, see here)
- currently unlimited storage capacity and no restrictions on individual file size
- supports private and public repositories
- publicly funded by the Federal Ministry of Education and Research (BMBF; details here)
- servers on German land (Munich, Germany; cf. GDPR)
- provides Digital Object Identifiers (DOIs) (details here) and allows free licensing (details here)
Sharing DataLad datasets via GitLab
“GitLab is open source software to collaborate on code. Manage git repositories with fine-grained access controls that keep your code secure.”
GitLab for Max Planck employees
- hosted by GWDG: gitlab.gwdg.de
- hosted by your institute, e.g., git.mpib-berlin.mpg.de
✨ Features of GitLab ✨
- free to use and open-source
- several MPS instances available (see above)
- supports private and public repositories
- use project management infrastructure (merge requests, issue boards, etc.) for your dataset projects
Publish and consume datasets like source code
Datasets can comfortably live in multiple locations:
Publication dependencies automate update in all places:
Redundancy: DataLad gets data from available sources
Clone the dataset from GitLab:
Access to special remotes needs to be configured:
DataLad retrieves data from available sources (here, GIN):
Summary
Summary and discussion
Science is complex
- Scientific units are not static: We need version control
- Science is modular: We need to link modular datasets
- Science is iterative: We need to establish provenance
- Science is collaborative and distributed: We want to share our work and integrate with diverse infrastructure
DataLad: Decentralized management of digital objects for open science
- DataLad can version control arbitrary datasets
- DataLad links modular version-controlled datasets
- DataLad establishes provenance and transparency
- DataLad integrates with diverse infrastructure
Develop everything like source code
- Code and data management using Git and DataLad (free, open-source command-line tools)
- Code and data sharing via flexible repository hosting services (GitLab, GitHub, GIN, etc.)
- Code and data storage on various infrastructure (GIN, OSF, S3, Keeper, Dataverse, and many more!)
- Project-related communication (ideas, problems, discussions) via issue boards on GitLab / GitHub etc.
- Transparent contributions to code and data via merge requests on GitLab (i.e., pull requests on GitHub)
- Reproducible procedures using datalad run, rerun, and containers-run commands (also Make etc.)
- Reproducible computational environments using software containers (e.g., Docker, Apptainer, etc.)
Overview of learning resources
Learn Git
- “Pro Git” by Scott Chacon & Ben Straub
- “Happy Git and GitHub for the useR” by Jenny Bryan, the STAT 545 TAs & Jim Hester
- “Version Control” by The Turing Way
- “Version Control with Git” by The Software Carpentries
- “Version control” (chapter 3 of “Neuroimaging and Data Science”) by Ariel Rokem & Tal Yarkoni
Learn DataLad
- “Datalad Handbook” by the DataLad team / Wagner et al., 2022, Zenodo
- “Research Data Management with DataLad” | Recording of a full-day workshop on YouTube
- Datalad on YouTube | Recorded workshops, tutorials and talks on DataLad
Learn both (disclaimer: shameless plug 🙈)
Full-semester course on “Version control of code and data using Git and DataLad” (v2.0!) in summer semester 2024 at University of Hamburg (generously funded by the Digital and Data Literacy in Teaching Lab program) with many open educational resources (online guide, quizzes and exercises)
References
Thank you!

Dr. Lennart Wittkuhn
wittkuhn@mpib-berlin.mpg.de
https://lennartwittkuhn.com/
Mastodon GitHub LinkedIn



Slides: https://lennartwittkuhn.com/talk-mps-fdm-2024
Source: https://github.com/lnnrtwttkhn/talk-mps-fdm-2024
Software: Reproducible slides built with Quarto and deployed to GitHub Pages using GitHub Actions for continuous integration & deployment
License: Creative Commons Attribution-ShareAlike 4.0 (CC BY-SA 4.0)
Contact: Feedback or suggestions via email or GitHub issues. Thank you!
Appendix
Example: “Let me just quickly copy those files …”
Without datalad run
Researcher shares analysis with collaborators.
With datalad run
Walkthrough: Sharing DataLad datasets via Keeper
Configure rclone:
rclone config
2024/03/19 11:45:32 NOTICE: Config file "/root/.config/rclone/rclone.conf" not found - using defaults
No remotes found, make a new one?
n) New remote
s) Set configuration password
q) Quit config
name> neuro-data
Option Storage.
Type of storage to configure.
Choose a number from below, or type in your own value.
1 / 1Fichier
\ (fichier)
2 / Akamai NetStorage
\ (netstorage)
3 / Alias for an existing remote
\ (alias)
4 / Amazon S3 Compliant Storage Providers including AWS, Alibaba, ArvanCloud, Ceph, ChinaMobile, Cloudflare, DigitalOcean, Dreamhost, GCS, HuaweiOBS, IBMCOS, IDrive, IONOS, LyveCloud, Leviia, Liara, Linode, Minio, Netease, Petabox, RackCorp, Rclone, Scaleway, SeaweedFS, StackPath, Storj, Synology, TencentCOS, Wasabi, Qiniu and others
\ (s3)
5 / Backblaze B2
\ (b2)
6 / Better checksums for other remotes
\ (hasher)
7 / Box
\ (box)
8 / Cache a remote
\ (cache)
9 / Citrix Sharefile
\ (sharefile)
10 / Combine several remotes into one
\ (combine)
11 / Compress a remote
\ (compress)
12 / Dropbox
\ (dropbox)
13 / Encrypt/Decrypt a remote
\ (crypt)
14 / Enterprise File Fabric
\ (filefabric)
15 / FTP
\ (ftp)
16 / Google Cloud Storage (this is not Google Drive)
\ (google cloud storage)
17 / Google Drive
\ (drive)
18 / Google Photos
\ (google photos)
19 / HTTP
\ (http)
20 / Hadoop distributed file system
\ (hdfs)
21 / HiDrive
\ (hidrive)
22 / ImageKit.io
\ (imagekit)
23 / In memory object storage system.
\ (memory)
24 / Internet Archive
\ (internetarchive)
25 / Jottacloud
\ (jottacloud)
26 / Koofr, Digi Storage and other Koofr-compatible storage providers
\ (koofr)
27 / Linkbox
\ (linkbox)
28 / Local Disk
\ (local)
29 / Mail.ru Cloud
\ (mailru)
30 / Mega
\ (mega)
31 / Microsoft Azure Blob Storage
\ (azureblob)
32 / Microsoft Azure Files
\ (azurefiles)
33 / Microsoft OneDrive
\ (onedrive)
34 / OpenDrive
\ (opendrive)
35 / OpenStack Swift (Rackspace Cloud Files, Blomp Cloud Storage, Memset Memstore, OVH)
\ (swift)
36 / Oracle Cloud Infrastructure Object Storage
\ (oracleobjectstorage)
37 / Pcloud
\ (pcloud)
38 / PikPak
\ (pikpak)
39 / Proton Drive
\ (protondrive)
40 / Put.io
\ (putio)
41 / QingCloud Object Storage
\ (qingstor)
42 / Quatrix by Maytech
\ (quatrix)
43 / SMB / CIFS
\ (smb)
44 / SSH/SFTP
\ (sftp)
45 / Sia Decentralized Cloud
\ (sia)
46 / Storj Decentralized Cloud Storage
\ (storj)
47 / Sugarsync
\ (sugarsync)
48 / Transparently chunk/split large files
\ (chunker)
49 / Union merges the contents of several upstream fs
\ (union)
50 / Uptobox
\ (uptobox)
51 / WebDAV
\ (webdav)
52 / Yandex Disk
\ (yandex)
53 / Zoho
\ (zoho)
54 / premiumize.me
\ (premiumizeme)
55 / seafile
\ (seafile)
Storage> seafile
Option url.
URL of seafile host to connect to.
Choose a number from below, or type in your own value.
1 / Connect to cloud.seafile.com.
\ (https://cloud.seafile.com/)
url> https://keeper.mpdl.mpg.de/
Option user.
User name (usually email address).
Enter a value.
user> wittkuhn@mpib-berlin.mpg.de
Option pass.
Password.
Choose an alternative below. Press Enter for the default (n).
y) Yes, type in my own password
g) Generate random password
n) No, leave this optional password blank (default)
y/g/n> y
Enter the password:
password:
Confirm the password:
password:
Option 2fa.
Two-factor authentication ('true' if the account has 2FA enabled).
Enter a boolean value (true or false). Press Enter for the default (false).
2fa> false
Option library.
Name of the library.
Leave blank to access all non-encrypted libraries.
Enter a value. Press Enter to leave empty.
library> neuro-data
Option library_key.
Library password (for encrypted libraries only).
Leave blank if you pass it through the command line.
Choose an alternative below. Press Enter for the default (n).
y) Yes, type in my own password
g) Generate random password
n) No, leave this optional password blank (default)
y/g/n> n
Edit advanced config?
y) Yes
n) No (default)
y/n> n
Configuration complete.
Options:
- type: seafile
- url: https://keeper.mpdl.mpg.de/
- user: wittkuhn@mpib-berlin.mpg.de
- pass: *** ENCRYPTED ***
- library: neuro-data
Keep this "neuro-data" remote?
y) Yes this is OK (default)
e) Edit this remote
d) Delete this remote
y/e/d> y
Current remotes:
Name Type
==== ====
neuro-data seafile
e) Edit existing remote
n) New remote
d) Delete remote
r) Rename remote
c) Copy remote
s) Set configuration password
q) Quit config
e/n/d/r/c/s/q> qdatalad push --to keeper
copy(ok): CHANGES (file) [to keeper...]
copy(ok): README (file) [to keeper...]
copy(ok): dataset_description.json (file) [to keeper...]
copy(ok): sub-01/anat/sub-01_T1w.nii (file) [to keeper...]
copy(ok): sub-01/func/sub-01_task-auditory_bold.nii (file) [to keeper...]
copy(ok): sub-01/func/sub-01_task-auditory_events.tsv (file) [to keeper...]
copy(ok): task-auditory_bold.json (file) [to keeper...] action summary:
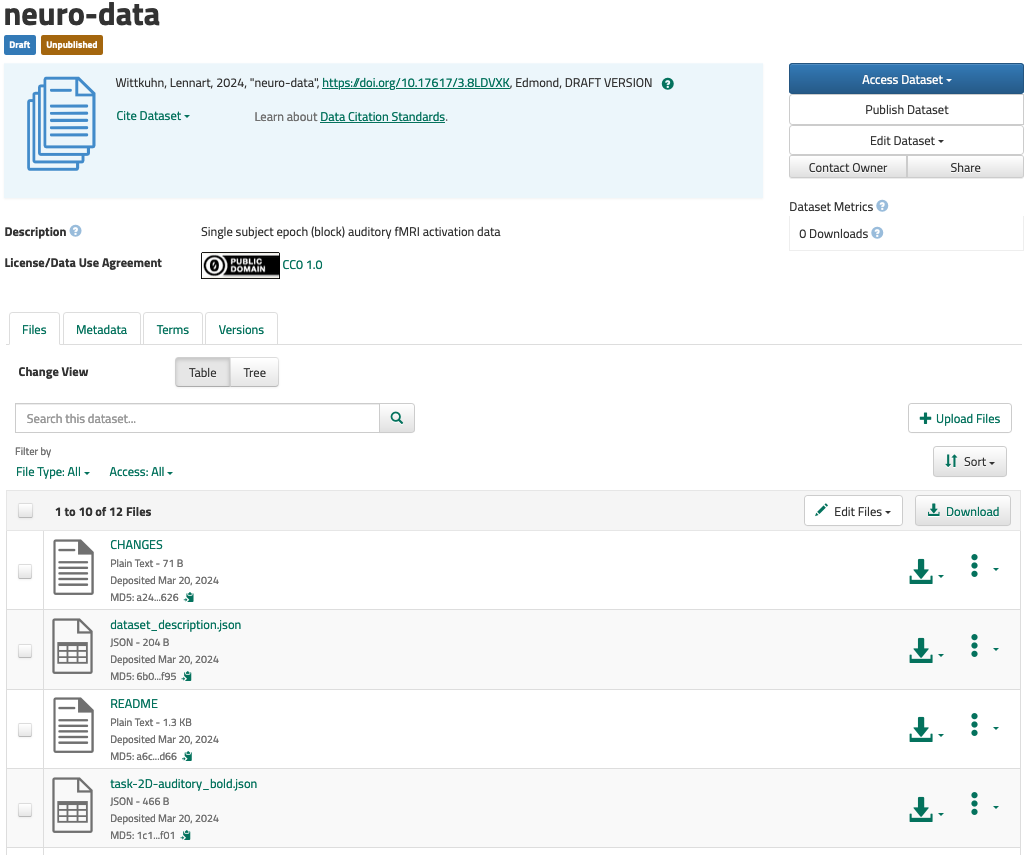
copy (ok: 7)Walkthrough: Sharing DataLad datasets via Edmond
If you want to publish a dataset to Dataverse, you will need a dedicated location on Dataverse that we will publish our dataset to. For this, we will use a Dataverse dataset3.
- Go to Edmond, log in, and create a new draft Dataverse dataset via the
Add Dataheader - The
New Datasetbutton takes you to a configurator for your Dataverse dataset. Provide all relevant details and metadata entries in the form4. Importantly, don’t upload any of your data files - this will be done by DataLad later. - Once you have clicked
Save Dataset, you’ll have a draft Dataverse dataset. It already has a DOI, and you can find it under the Metadata tab as “Persistent identifier”: - Finally, make a note of the URL of your dataverse instance (e.g., https://edmond.mpg.de/), and the DOI of your draft dataset. You will need this information for step 3.
Add a Dataverse sibling to your dataset
We will use the datalad add-sibling-dataverse command. This command registers the remote Dataverse Dataset as a known remote location to your Dataset and will allow you to publish the entire Dataset (Git history and annexed data) or parts of it to Dataverse.
If you run this command for the first time, you will need to provide an API Token to authenticate against the chosen Dataverse instance in an interactive prompt. This is how this would look:
A dataverse API token is required for access. Find it at https://edmond.mpg.de by clicking on your name at the top right corner and then clicking on API Token
token:
A dataverse API token is required for access. Find it at https://edmond.mpg.de by clicking on your name at the top right corner and then clicking on API Token
token (repeat):
Enter a name to save the credential securely for future reuse, or 'skip' to not save the credential
name: You’ll find this token if you follow the instructions in the prompt under your user account on your Dataverse instance, and you can copy-paste it into the command line.
A dataverse API token is required for access. Find it at https://edmond.mpg.de by clicking on your name at the top right corner and then clicking on API Token
token:
A dataverse API token is required for access. Find it at https://edmond.mpg.de by clicking on your name at the top right corner and then clicking on API Token
token (repeat):
Enter a name to save the credential securely for future reuse, or 'skip' to not save the credential
name: skipAs soon as you’ve created the sibling, you can push:
copy(ok): CHANGES (file) [to dataverse-storage...]
copy(ok): README (file) [to dataverse-storage...]
copy(ok): dataset_description.json (file) [to dataverse-storage...]
copy(ok): sub-01/anat/sub-01_T1w.nii (file) [to dataverse-storage...]
copy(ok): sub-01/func/sub-01_task-auditory_bold.nii (file) [to dataverse-storage...]
copy(ok): sub-01/func/sub-01_task-auditory_events.tsv (file) [to dataverse-storage...]
copy(ok): task-auditory_bold.json (file) [to dataverse-storage...]
publish(ok): . (dataset) [refs/heads/main->dataverse:refs/heads/main [new branch]]
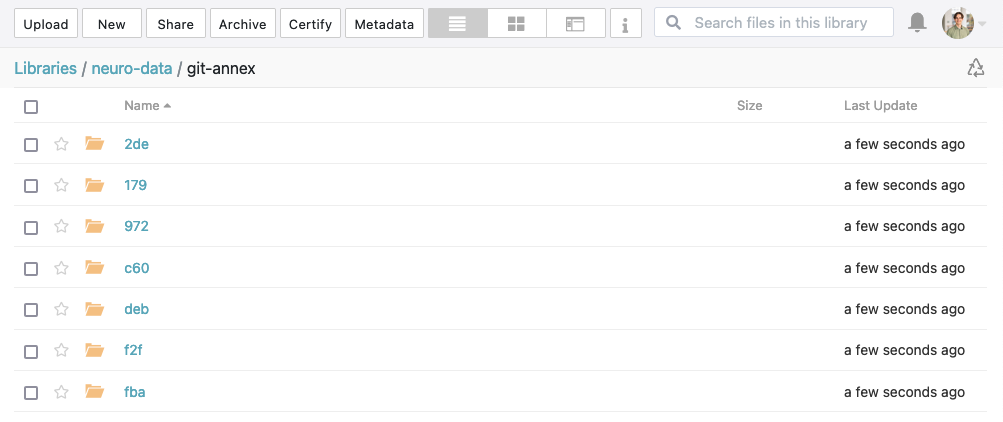
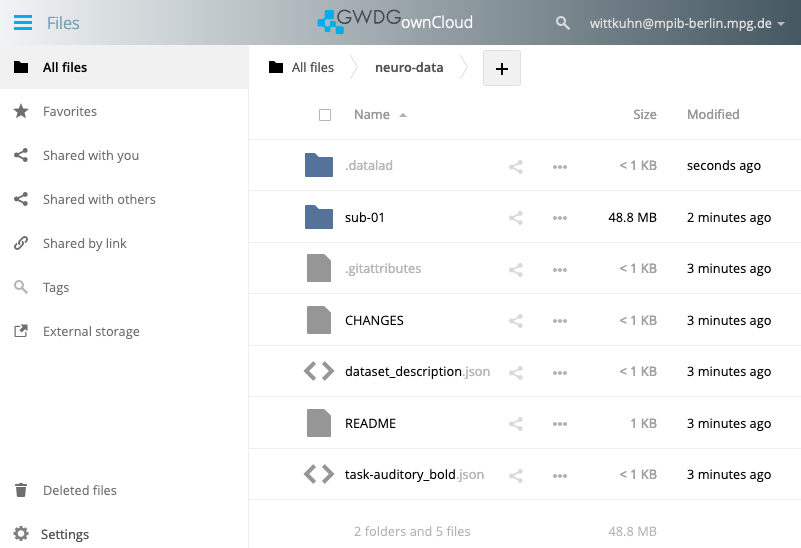
publish(ok): . (dataset) [refs/heads/git-annex->dataverse:refs/heads/git-annex [new branch]] Walkthrough: Sharing DataLad datasets via ownCloud / nextCloud
Get the WebDAV address
- Click on
Settings(bottom left) - Copy the WebDAV address, for example:
https://owncloud.gwdg.de/remote.php/nonshib-webdav/
datalad create-sibling-webdav \
--dataset . \
--name owncloud-gwdg \
--mode filetree \
1 'https://owncloud.gwdg.de/remote.php/nonshib-webdav/<dataset-name>'- 1
-
Replace
<dataset-name>with the name of your dataset, i.e., the name of your dataset folder. In this example, we replace<dataset-name>withneuro-data. The complete command for your example hence looks like this:
You will be asked to provide your ownCloud account credentials:
- 1
- Enter the email address of your ownCloud account.
- 2
- Enter the password of your ownCloud account.
- 3
- Repeat the password of your ownCloud account.
create_sibling_webdav.storage(ok): . [owncloud-gwdg-storage: https://owncloud.gwdg.de/remote.php/nonshib-webdav/neuro-data]
[INFO ] Configure additional publication dependency on "owncloud-gwdg-storage"
create_sibling_webdav(ok): . [owncloud-gwdg: datalad-annex::?type=webdav&encryption=none&exporttree=yes&url=https%3A//owncloud.gwdg.de/remote.php/nonshib-webdav/neuro-data]Finally, we can push the dataset to ownCloud:
1datalad push --to owncloud-gwdg- 1
-
Use
datalad pushto push the dataset contents to ownCloud. For details ondatalad push, see the command line reference and this chapter in the DataLad Handbook.
We can now view the files on ownCloud and inspect them through the web browser.
Reproducible Research Data Management with DataLad
Footnotes
(Source: Wikipedia)
see DataLad dataset of 80TB / 15 million files from the Human Connectome Project (see details)
Dataverse datasets contain digital files (research data, code, …), amended with additional metadata. They typically live inside of dataverse collections.
At least,
Title,Description, andOrganizationare required.